Table of Contents
Research Overview
We are a computational genetics and genomics lab. Our main research is on genetic variation, including mechanisms of spontaneous mutagenesis, functional effects of mutations and allelic variants, population genetics and the relationship between genotype and phenotype. As part of our research we develop new computational and statistical methods to assist DNA sequencing studies. Below, we list our research areas through recent publications (some publications belong to more than one area). All publications by the lab can be found here.
Mutagenesis
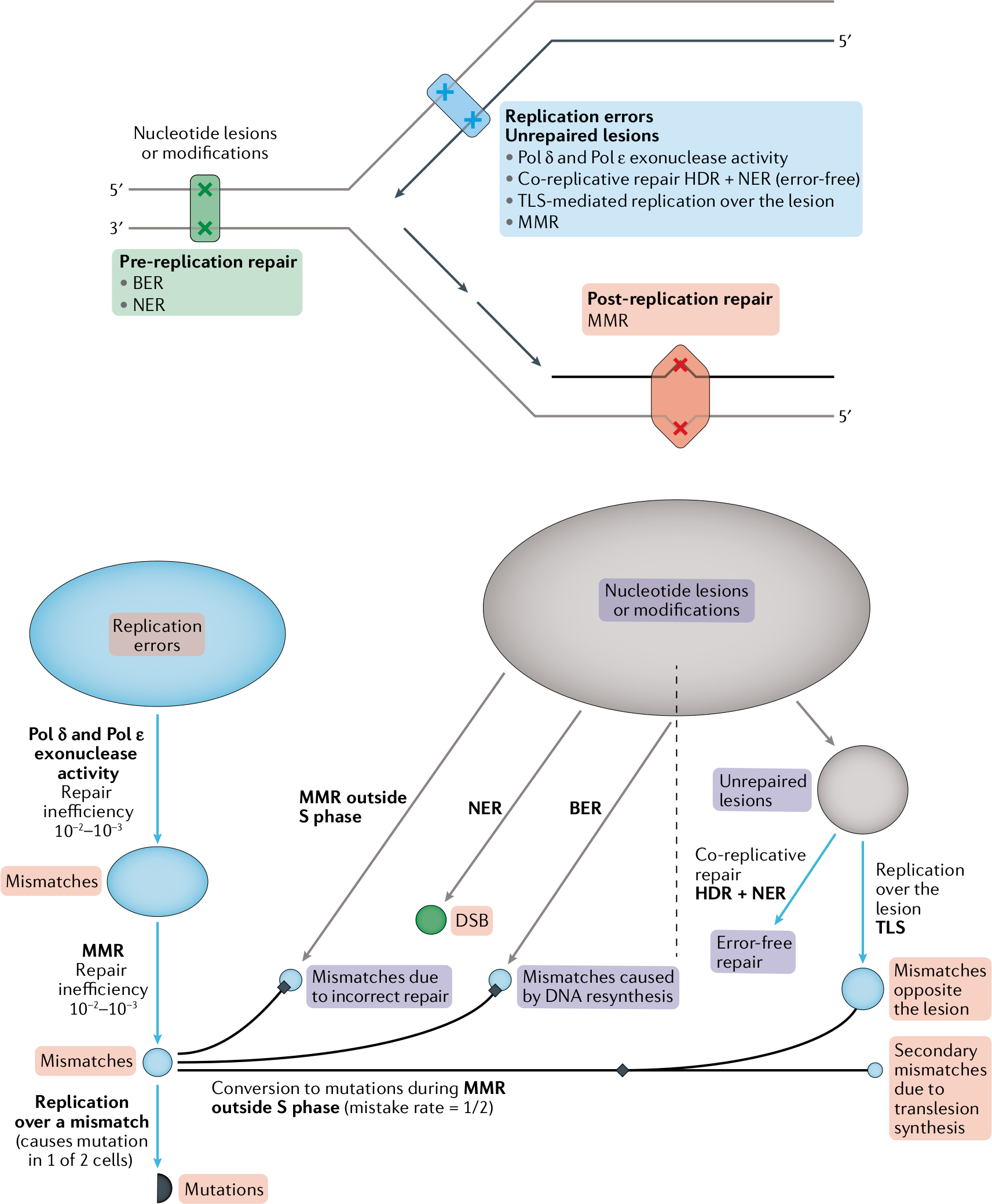
Mutations are the source of population genetic variation; they fuel evolution and cause disease. Studies of mutagenesis shed light on mechanisms of the genome maintenance including DNA replication and repair. Data on de novo germ-line mutations are now available from whole genome sequencing of parent-child trios. There is also a growing amount of data on somatic mutations in both cancerous and healthy tissues We analyze statistical properties of mutations alongside epigenomic datasets. We believe that this analysis has a potential to generate biologically relevant hypotheses on leading mechanisms of spontaneous mutations in humans. From an evolutionary viewpoint, it can be informative about the evolution of mutation rate. On the practical side, accurate models of mutation rate will enhance statistical methods aimed at mapping genes using recurrent de novo mutations. By statistically decomposing mutation rate variation along the genome, we characterized several mutagenic processes. We demonstrated the impact of bulky DNA damage on mutagenesis in germ-line. We showed that enzymatic demethylation and transcription by polymerase III are mutagenic. We identified genomic regions of high mutability in non-dividing oocytes. We have also developed a base-pair resolution map of mutation rate for the human genome. For somatic cancer mutations, we demonstrated that the relationship between chromatin accessibility and modification and mutation rate is highly cell-type specific.
Population genetics
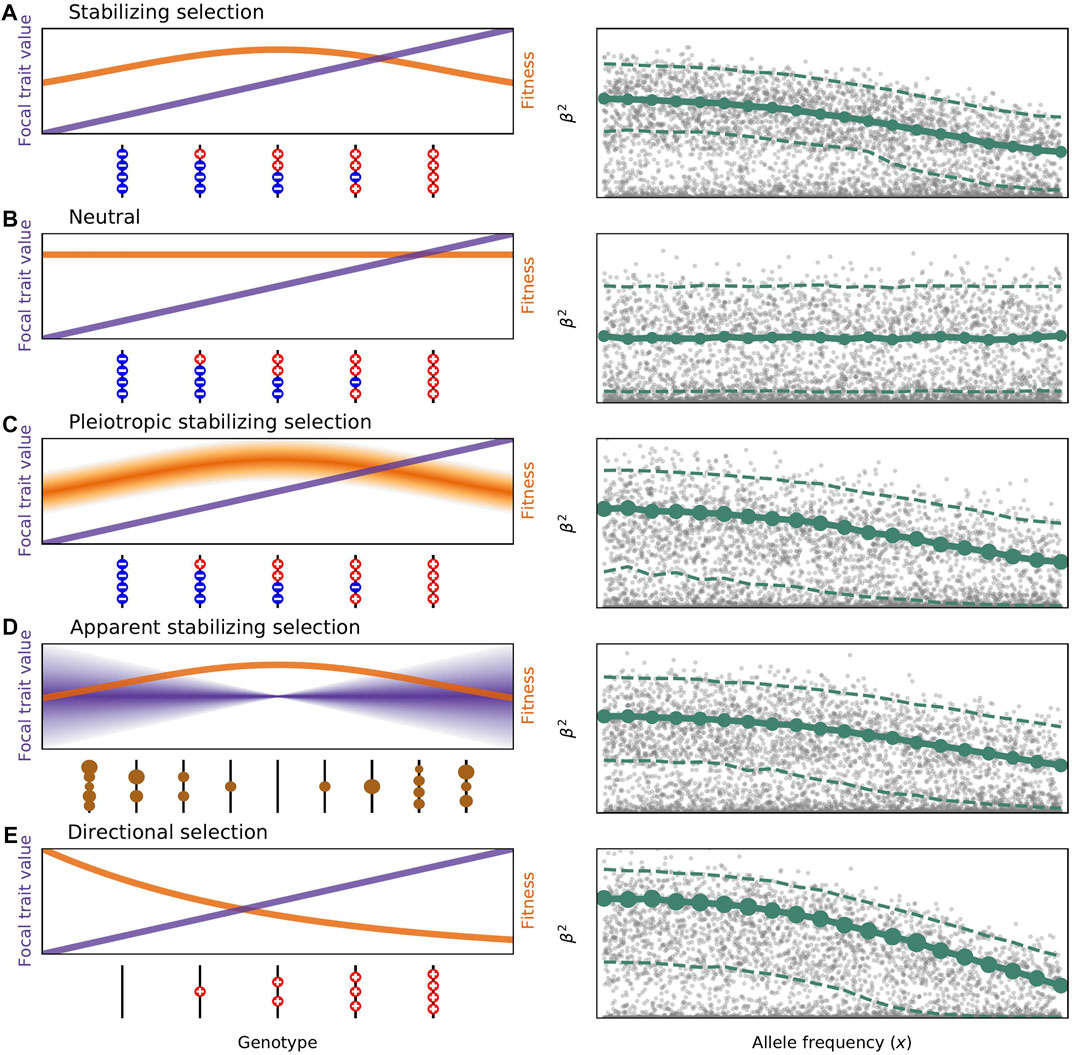
We are interested in population genetics as a lens through which we can study microevolution. Dynamics of allele propagation in populations depends on a number of evolutionary forces. Now, development of theoretical models is enhanced by the availability of massive sequencing datasets. Our results include the demonstration that deleterious alleles are younger than neutral alleles at the same population frequency. We studied the effect of population bottlenecks and expansions on the burden of deleterious mutations under arbitrary dominance coefficient. We are broadly interested in estimating of the strength of natural selection acting on human deleterious variants and epistatic interactions. We are working on maps of selective constraints in coding and non-coding fractions of the human genome. A separate direction of research is on mechanisms of maintenance of phenotypic variation. We are testing predictions of classical population genetics models using the vast trove of GWAS results on multiple human phenotypes.
Statistical genetics of complex traits and effect of allelic variants
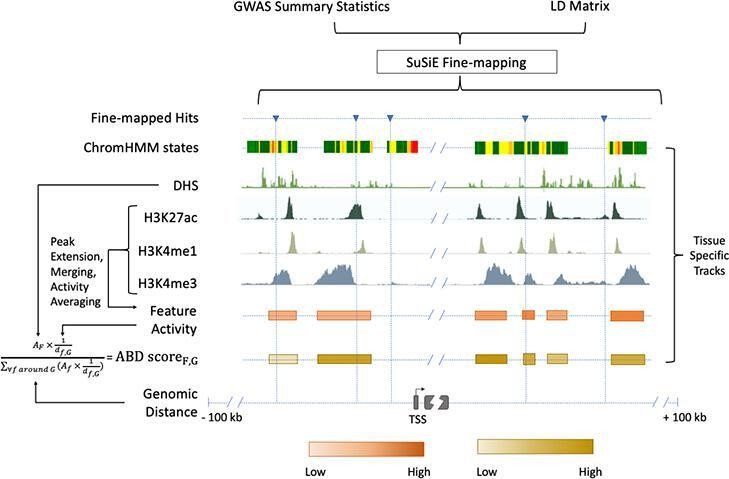
Massive efforts in complex trait genetics resulted in the identification of numerous allelic variants in phenotyped individuals. However, the functional effects of genetic variants at the molecular level are poorly understood. The field is also lacking understanding of the effect of genetic variation on phenotypes from the perspective of larger biological units such as pathways and networks. We have been working on predicting and interpreting the effect of allelic variants in coding (e.g. PolyPhen2) and noncoding variants. We showed that most genetic changes affecting baseline expression of well-established disease genes do not result in the phenotypic change. Conversely, only a minority of known association signals are likely explained by changes in bulk expression. We are now exploring the effect of context on the function of regulatory variants. To study the effect of allelic variation on complex phenotypes at the higher organizational level, we are developing rare variant association tests that are informed by biological knowledge at the level of pathways and networks.
Applications to cancer genomics and genetics of rare diseases
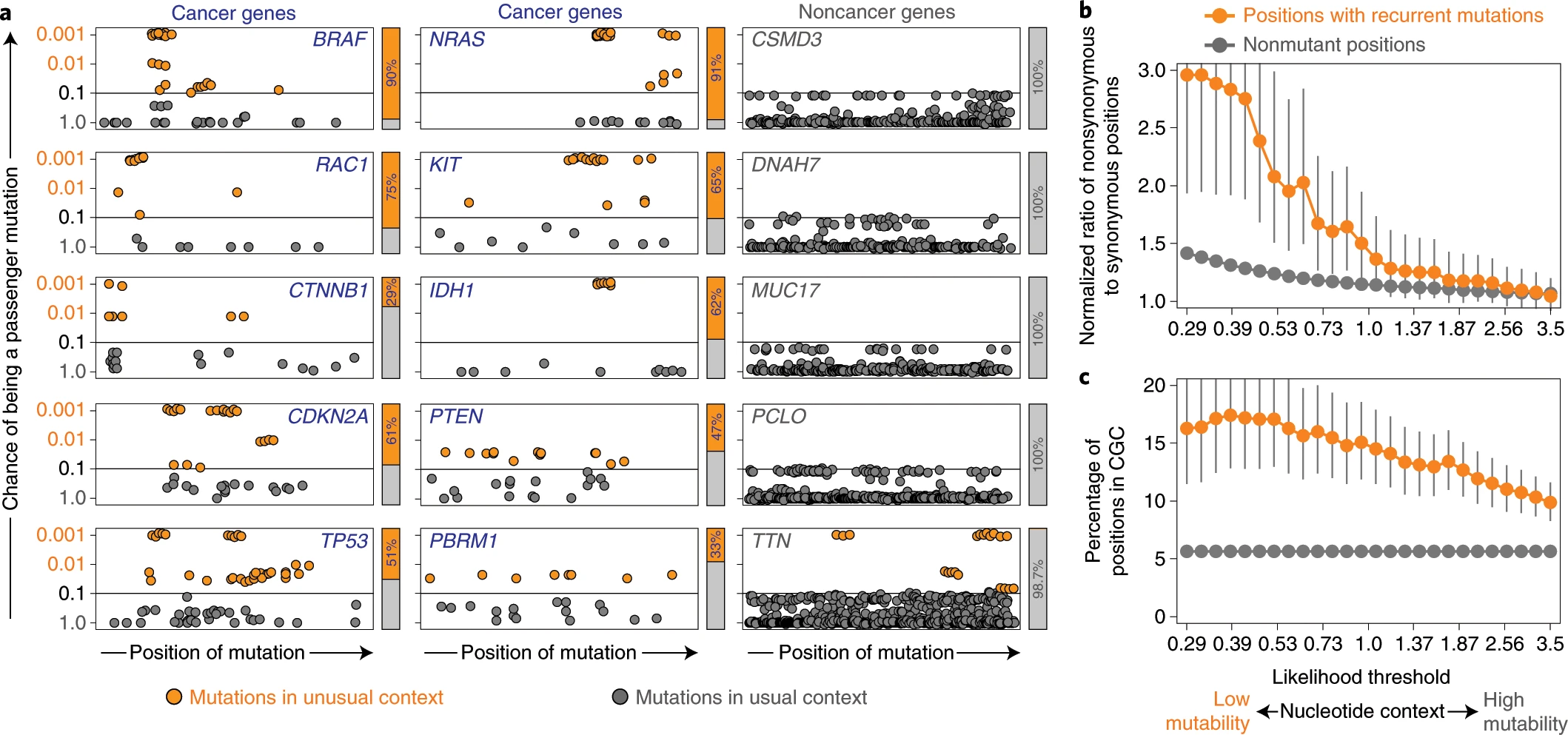
The lab is actively involved in Mendelian disease genomic research and participates in the Undiagnosed Diseases Network (UDN), a research study backed by the NIH Common Fund program. With the accumulation of genetic and phenotypic data Mendelian genetics becomes amenable to population-level statistical approaches. The genome is a finite place, and in a sufficiently large dataset multiple individuals will possess mutations in the same gene. We are developing and applying new computational methods to cohorts of rare disease patients. We have also been developing methods for cancer genomics and analyzed cancer genomics datasets.